P2: Design and Characterization of Protein Modules for MC-Based Sensing and Control in Microliter-scale Bioreactors
 Motivation and state of the art:
Motivation and state of the art:
Miniaturization is crucial for accelerating the implementation and optimization of biotechnological processes1. One present research focus is the further miniaturization towards smaller working volumes (< 100 µL) while preserving high-precision reaction control. For that purpose, project P1 will develop a modular toolbox of nanodevices for monitoring and controlling reaction conditions in microliter-scale bioreactors. The design and implementation of such nanodevices is very challenging, which is particularly true for systems employing proteins as building blocks. We have previously identified a set of candidate proteins with favourable properties for MC-based sensing and control2; however, the design of functional synthetic nanodevices will in most cases require tailoring of protein properties to the particular needs of the system. Rational protein design is a powerful approach for the modulation of protein properties by site-directed mutagenesis. This approach has already been successfully applied to alter binding properties, catalytic properties, toxicity, or immunogenicity3-7. Up to now, however, only few proteins have been systematically optimized for the inclusion in synthetic nanodevices. Consequently, there is a need for a defined set of proteins that can be used for different molecular tasks in such devices.
Objectives:
This project aims to develop a modular toolbox of protein components that can be used in nanodevices for monitoring and controlling reaction conditions in microliter-scale bioreactors. Particular focus will be on proteins that can be used as components of the nanodevices developed in P1 to enhance their versatility. For that purpose, we will investigate three classes of modules with different scopes of application:
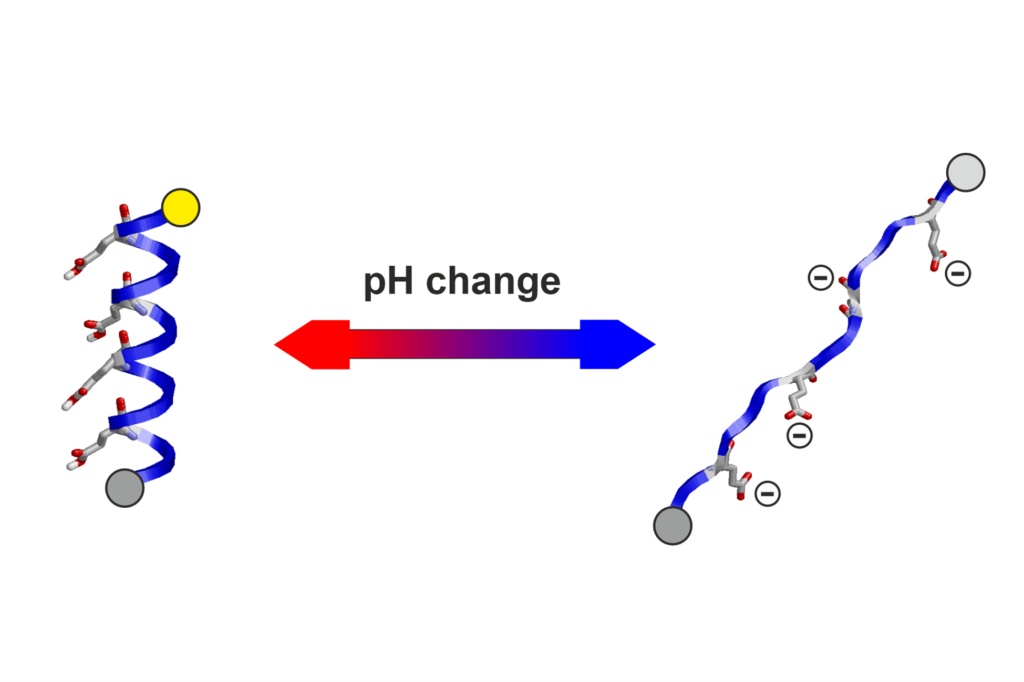
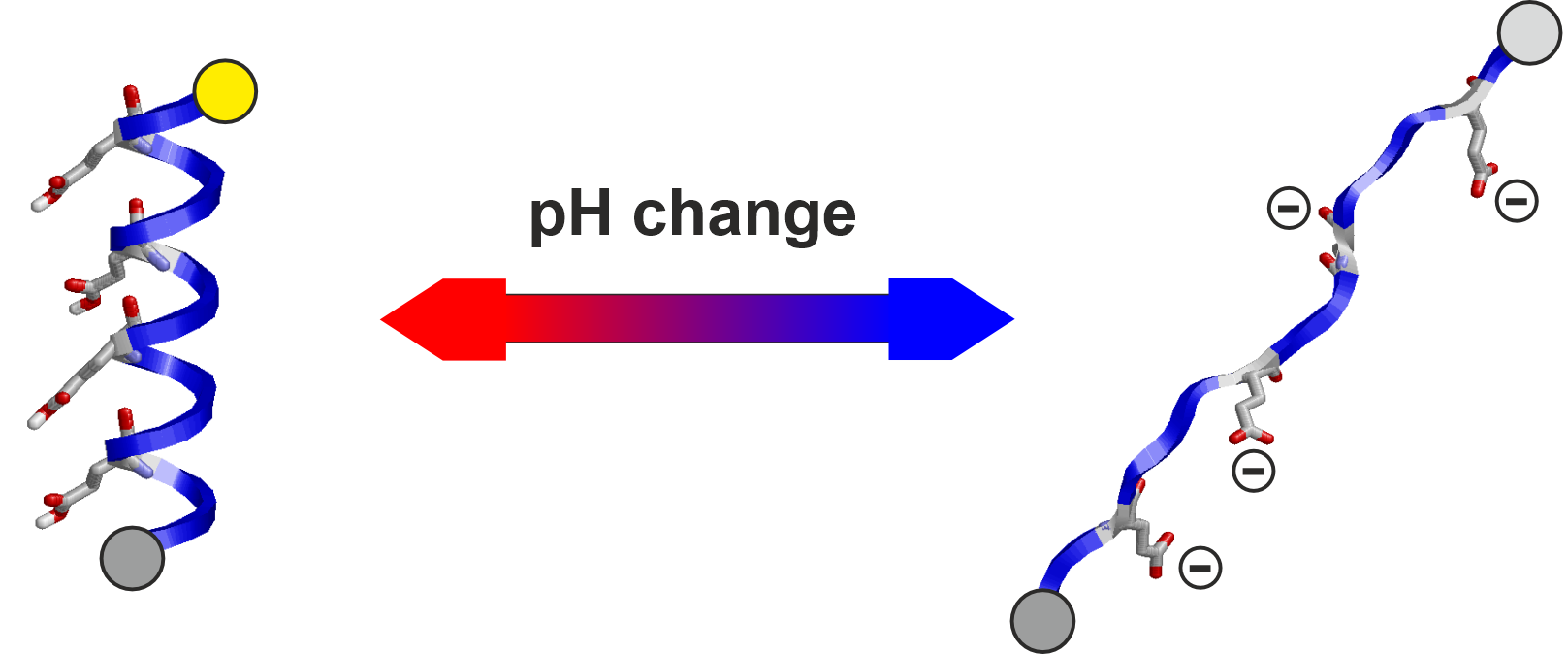
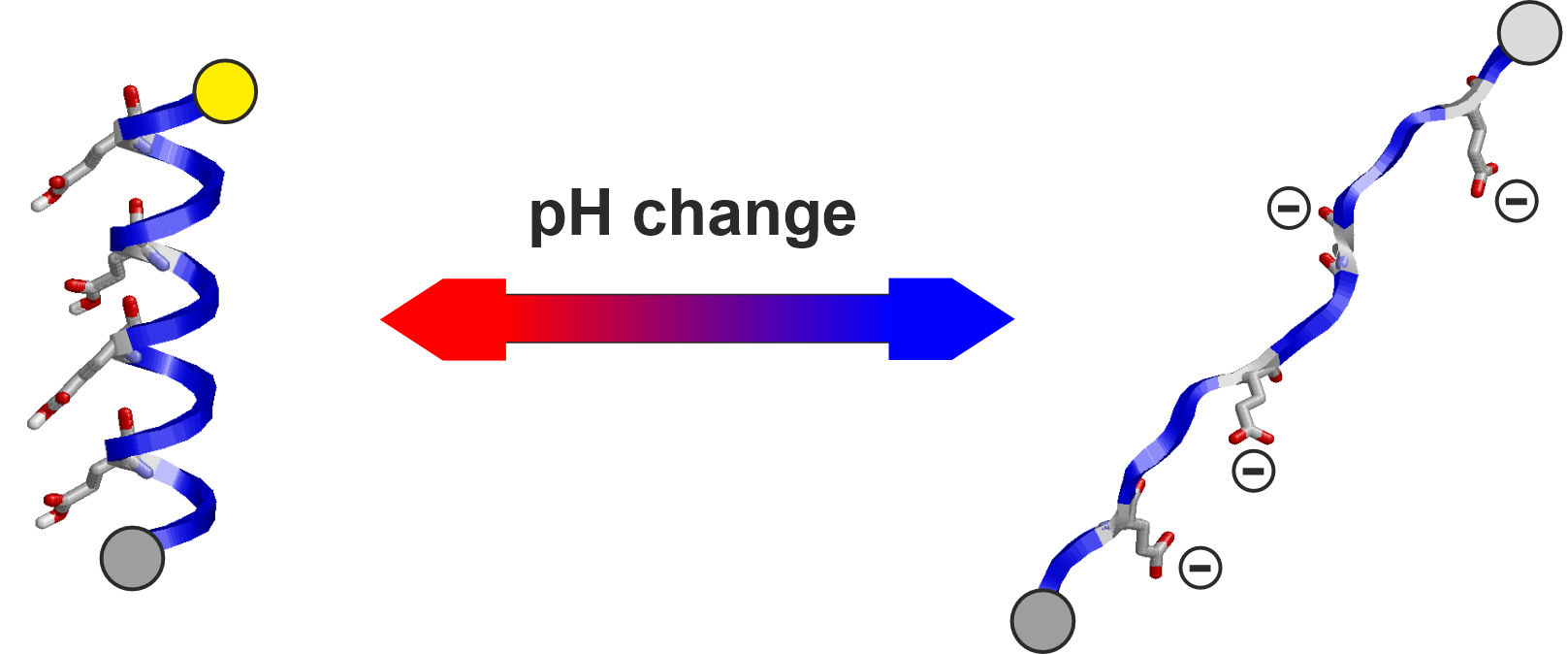
- pH-detection modules,
- substrate-detection modules,
- substrate-exchange modules.
For each class of modules, candidate proteins will be identified and subsequently modified to obtain favourable molecular properties for the inclusion in MC-based sensing and control systems. The impact of the investigated protein modules on the overall properties of the nanodevices developed in P1 will be assessed by MC-based modelling in P3.
References
1.Ladner, A. Grünberger, C. Probst, D. Kohlheyer, J. Büchs, and F. Delvigne. “15- Application of Mini- and Micro-Bioreactors for Microbial Bioprocesses,“ in Current Develop. Biotechnol. Bioeng., Eds. C. Larroche, M. Sanromán, G. Du, and A. Pandey (Amsterdam: Elsevier), pp. 433–461, 2017, doi: 10.1016/B978-0-444-63663-8.00015-X.
2.Söldner, E. Socher, V. Jamali, W. Wicke, A. Ahmadzadeh, H. Breitinger, A. Burkovski, K. Castiglione, R. Schober, and H. Sticht. “A Survey of Biological Building Blocks for Synthetic Molecular Communication Systems”. IEEE Commun. Surv. Tutor., vol. 22, pp. 2765-2800, Fourthquarter 2020.
3.Jiang, E. Althoff, F. Clemente, L. Doyle, D. Rothlisberger, A. Zanghellini, J. Gallaher, J. L. Betker, F. Tanaka, C. Barbas, 3rd, D. Hilvert, K. Houk, B. Stoddard, and D. Baker, „De Novo Computational Design of Retro-aldol Enzymes,“ Science, vol. 319, pp. 1387-91, Mar. 2008.
4.Tournier, C. Topham, A. Gilles, B. David, C. Folgoas, E. Moya-Leclair, E. Kamionka, M. Desrousseaux, H. Texier, S. Gavalda, M. Cot, E. Guemard, M. Dalibey, J. Nomme, G. Cioci, S. Barbe, M. Chateau, I. Andre, S. Duquesne, and A. Marty, „An Engineered PET Depolymerase to Break Down and Recycle Plastic Bottles,“ Nature, vol. 580, pp. 216-219, Apr. 2020.
5.Lechner, N. Ferruz, and B. Hocker, „Strategies for Designing Non-natural Enzymes and Binders,“ Curr. Opin. Chem. Biol., vol. 47, pp. 67-76, Dec. 2018.
6.Müller-Schiffmann, A. Andreyeva, A. H. Horn, K. Gottmann, C. Korth, and H. Sticht, „Molecular Engineering of a Secreted, Highly Homogeneous, and Neurotoxic Abeta Dimer,“ ACS Chem. Neurosci., vol. 2, pp. 242-8, May 2011.
7.Sesterhenn, C. Yang, J. Bonet, J. Cramer, X. Wen, Y. Wang, C. Chiang, L. Abriata, I. Kucharska, G. Castoro, S. S. Vollers, M. Galloux, E. Dheilly, S. Rosset, P. Corthesy, S. Georgeon, M. Villard, C. Richard, D. Descamps, T. Delgado, E. Oricchio, M. A. Rameix-Welti, V. Mas, S. Ervin, J. Eleouet, S. Riffault, J. Bates, J. Julien, Y. Li, T. Jardetzky, T. Krey, and B. Correia, „De Novo Protein Design Enables the Precise Induction of RSV-neutralizing Antibodies,“ Science, vol. 368, p. eaay5051, May 2020.